Significance
Transmembrane (TM) domains are regions within membrane proteins that span across the lipid bilayer of cell membranes. These domains are predominantly hydrophobic, allowing them to interact with the hydrophobic core of the lipid bilayer, and they play a crucial role in the structure and function of membrane proteins. Membrane proteins, including those with TM domains, are essential for various cellular processes, such as signal transduction, substance transport, and cell-cell communication. TM domains can be composed of α-helices, β-barrels, or other structures, depending on the protein. The α-helical TM domains are more common in eukaryotic cells, while β-barrel TM domains are found in the outer membranes of gram-negative bacteria, mitochondria, and chloroplasts. Targeting the TM regions of membrane proteins can be challenging due to their hydrophobic nature and their location within the cell membrane. However, several strategies have been developed for this purpose. For instance, small hydrophobic molecules can be designed to penetrate the cell membrane and interact with the TM domains of membrane proteins, modulating their activity. This approach is commonly used in drug development. Moreover, while it’s challenging to target TM domains with traditional antibodies due to their hydrophobicity and inaccessibility, engineered antibody fragments or single-domain antibodies (nanobodies) have been developed to target extracellular regions adjacent to TM domains or exposed epitopes of the TM domains themselves. Additionally, peptides that mimic the structure or function of TM domains can be used to interfere with the protein-protein interactions involving TM domains. These peptides can either be cell-penetrating or designed to interact with extracellular parts of the TM domains. Each of these strategies has its advantages and limitations, and the choice of strategy depends on the specific characteristics of the TM domain and the membrane protein being targeted. Advances in structural biology, molecular modeling, and biotechnology continue to improve the precision and efficacy of targeting membrane proteins’ TM regions for therapeutic purposes. The erythropoietin receptor (EPOR), a member of the cytokine receptor family, exemplifies a TM domain-containing protein that is pivotal in erythropoiesis. The erythropoietin receptor plays a pivotal role in this process by mediating the effects of erythropoietin (EPO), a glycoprotein hormone that is the primary regulator of erythropoiesis. EPOR is a member of the cytokine receptor superfamily and is primarily expressed on the surface of erythroid progenitor cells in the bone marrow. It is a transmembrane protein that, upon binding to erythropoietin, undergoes a conformational change that enables it to dimerize (pair with another EPOR molecule) or oligomerize (form complexes with multiple EPOR molecules). This structural change activates intracellular signaling pathways that are crucial for the survival, proliferation, and differentiation of erythroid progenitor cells into mature red blood cells. The erythropoietin receptor has been a focus of clinical interest due to its role in various disorders and its potential as a therapeutic target. For instance, in conditions such as chronic kidney disease, where the production of EPO by the kidneys is impaired, anemia can result due to insufficient erythropoiesis. Recombinant human erythropoietin and EPO-stimulating agents that target EPOR are used to treat anemia in CKD and other conditions. Ongoing research is focused on further understanding the signaling mechanisms of EPOR, the development of novel ESAs with improved safety and efficacy profiles, and exploring the potential of EPOR as a target for treating anemia of chronic disease and other disorders. Traditional strategies targeting membrane proteins have predominantly focused on their extracellular domains; however, the TM domains, embedded within the lipid bilayer, present a largely unexploited avenue for therapeutic intervention. The erythropoietin receptor remains a vital area of study in hematology, with significant implications for the treatment of anemia and beyond.
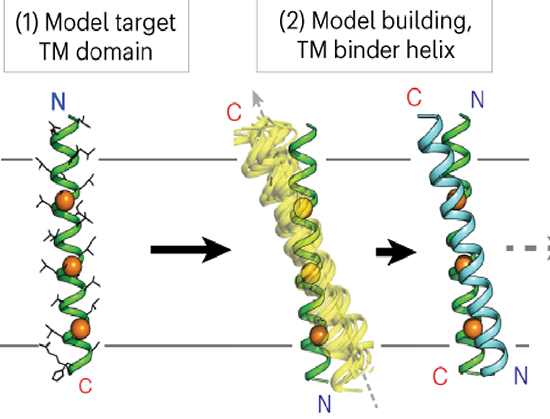
A new study published in Nature Chemical Biology and led by Professor Marco Mravic, Dr. Li He, Dr. Huong Kratochvil, Dr. Hailin Hu, Dr. Sarah Nick, Dr. Weiya Bai, Dr. Anne Edwards, Dr. Hyunil Jo, Dr. Yibing Wu, Professor Daniel DiMaio & Professor William DeGrado from Yale university and University of California San Francisco, the authors leveraged advanced computational algorithms to design synthetic TM domains. By focusing on the EpoR as a model system, they ingeniously encoded interactions in an antiparallel helix topology, distinct from the receptor’s native dimerization interface. This innovative strategy involved meticulous computational modeling, guided by structural informatics and biophysical principles, to predict and optimize the interactions between the designed TM protein and EpoR. Indeed, computational design is emerging as a formidable tool in the realm of protein engineering, offering the precision to tailor molecules that can specifically interact with target proteins at their TM domains. The authors demonstrated the de novo design of TM proteins that bind to the EpoR TM domain in a topology that competitively inhibits its dimerization and, consequently, its signaling. Such an approach circumvents the limitations of previous methods that were largely confined to mimicking natural interactions, paving the way for creating novel molecules with tailored functionalities.
The researchers showed that expression of these synthetic TM proteins in mammalian cells were able to complex with EpoR, effectively inhibiting its function. This finding not only validates the computational design approach but also opens new avenues for modulating EpoR activity in a controlled manner. Given EpoR’s critical role in erythropoiesis, the ability to influence its signaling pathway holds potential therapeutic value for conditions such as anemia and polycythemia. While this study marks a significant advance in targeting TM domains via computational design, it also highlights the challenges that lie ahead. The complexity of membrane protein structures and their dynamic nature in the lipid environment necessitates further refinement of computational models and experimental validation methods. Moreover, the translation of these findings into therapeutic applications will require a comprehensive understanding of the long-term effects of such interventions on cellular functions and organismal physiology. In conclusion, the authors successfully harnessed the power of computational design to target the TM domains of membrane proteins, specifically the erythropoietin receptor, this study broadens our understanding of protein-protein interactions within the membrane and also lays the groundwork for novel therapeutic strategies targeting a range of diseases.
Reference
Mravic M, He L, Kratochvil HT, Hu H, Nick SE, Bai W, Edwards A, Jo H, Wu Y, DiMaio D, DeGrado WF. De novo-designed transmembrane proteins bind and regulate a cytokine receptor. Nat Chem Biol. 2024. doi: 10.1038/s41589-024-01562-z.
 Medicine Innovates Medicine Innovates: Delivering innovations in medicine to the world for better health and prosperity
Medicine Innovates Medicine Innovates: Delivering innovations in medicine to the world for better health and prosperity


