Significance
The field of synthetic biology has long been driven by the desire to take control of the protein translation and make it most beneficial for biotechnology from mass production of industrial enzymes, therapeutic proteins, or the exploration of novel biosynthetic pathways. To do this, researchers depend on the ribosome’s capacity to efficiently build polypeptides from genetic instructions. However, despite decades of refinement in expression systems, protein synthesis remains unpredictable. Some sequences translate seamlessly in Escherichia coli, which is the main workhorse of biotechnology, while others stall, collapse in yield, or disappear into insoluble aggregates. This inconsistency represents a major bottleneck in biotechnology and industrial pipelines alike. One major challenge is sequence-dependent phenomena that slow or halt ribosomal progression. Codon bias, stable mRNA secondary structures, and the scarcity of particular tRNAs are familiar culprits. But in recent years attention has shifted to ribosome arrest peptides (RAPs)—short motifs within nascent chains that directly interfere with elongation. Unlike codon usage effects, which operate through nucleic acid features, RAPs act at the level of the growing peptide itself. As the emerging chain interacts with the ribosome exit tunnel, certain motifs can stall elongation in a highly sequence-specific manner. Well-studied examples include SecM in E. coli and MifM in Bacillus subtilis, each harnessed by the cell for regulatory purposes. For synthetic biologists, however, such motifs are obstacles, introducing unexpected translation pauses and sharply reducing yields.
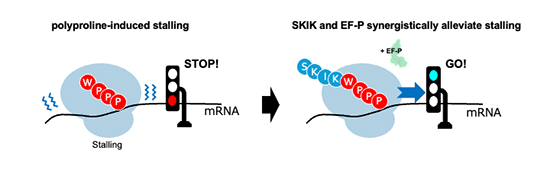
Among the most problematic motifs are those rich in consecutive prolines which is considered rigid structure of proline that slows peptide bond formation, and when multiple prolines align in series, the ribosome can grind to a halt. The artificial WPPP sequence, containing three consecutive prolines, exemplifies this challenge. Traditional workarounds—such as elongation factor P (EF-P), a specialized bacterial protein that alleviates polyproline stalling—are not always sufficient. Even with EF-P present, many constructs containing proline-rich regions remain difficult to express, leaving biologists searching for additional solutions. Recently, a surprising observation from Nagoya University discovered that the short SKIK peptide, which is a tetrapeptide of serine, lysine, isoleucine, and lysine, increase protein yields when fused to challenging targets. What began as a convenient tag for expression unexpectedly revealed itself as more than a passive handle: the nascent SKIK sequence itself appeared to alter the ribosome’s behavior. In earlier work, Ojima-Kato and colleagues showed that SKIK could even counteract stalling by the SecM arrest peptide. These findings raised a provocative possibility—that translation could be actively promoted by carefully positioned nascent sequences, rather than only impeded by them.
To this account, new research paper published in ACS Synthetic Biology and conducted by Yuma Nishikawa, Riko Fujikawa, Hideo Nakano, and led by Professor Teruyo Ojima-Kato from the Nagoya University alongside Dr. Takashi Kanamori from GeneFrontier Corporation in Japan, the researchers developed a simple yet powerful strategy to overcome ribosomal stalling by inserting a short SKIK tetrapeptide near polyproline motifs. Through both in vitro and in vivo experiments, they demonstrated that SKIK enhances translation efficiency, accelerates ribosome turnover, and synergizes with elongation factor P to maximize protein yield. Their approach transforms a tiny nascent peptide sequence into a functional tool for boosting recombinant protein production, offering a minimal and easily adoptable solution for synthetic biology. The researchers began with DNA constructs engineered to position the SKIK tetrapeptide at varying distances from the problematic WPPP motif. In a cell-free protein synthesis system based on purified components, they used superfolder GFP as a reporter to provide a clear, real-time measure of translation efficiency. Their idea was that if SKIK could counteract ribosomal arrest, then fluorescence should rise when it was placed in proximity to WPPP. They found that when SKIK was positioned immediately upstream of WPPP, protein production increased markedly compared to constructs where SKIK was farther away. Western blotting confirmed that full-length GFP accumulated in higher amounts under these conditions,
Afterward, the team substituted SKIK with alternative tetrapeptides such as GGGG, AAAA, LLLL, and IIII to ensure the result was not due to nucleotide composition. Despite similar GC contents in some cases, these replacements failed to match SKIK’s potency. Some offered modest relief, which suggest that multiple sequences can exert small influences, yet SKIK consistently outperformed them. The authors’ finding highlighted that the benefit was not simply structural randomness but encoded within the unique chemistry of SKIK itself. Moving into E. coli cultures, the researchers repeated the constructs using autoinduction media and found that strains carrying SKIK close to WPPP displayed visibly brighter fluorescence and stronger GFP bands, while controls lagged behind. The in vivo environment, rich with elongation factors and quality-control systems, seemed to amplify the effect, reinforcing the view that SKIK acted through a mechanism tightly coupled to ribosomal function during active translation.
Professor Teruyo Ojima-Kato and colleagues also monitored GFP production over time and fitted their data to a kinetic model inspired by enzyme catalysis (treating ribosomes as enzymes, mRNA as substrate, and protein as product) and they found that SKIK boosted the maximum translation rate more than twentyfold, and enhanced both initiation and elongation constants. In practical terms, ribosomes turned over more efficiently and completed proteins at a faster pace when SKIK was present. The precision of this modeling gave numerical weight to what had been qualitative observations, transforming the SKIK effect into a quantifiable kinetic phenomenon. Finally, they explored how SKIK interacted with EF-P, the known rescue factor for proline-rich stalls. When added together, SKIK and EF-P synergized, producing protein levels far above either alone. However, free SKIK peptide added externally to the reaction had no impact, which proved that the effect depended on SKIK being translated as part of the nascent chain.
In conclusion, until recently, arrest peptides were seen almost exclusively as molecular brakes, tools that cells evolved to regulate gene expression through controlled stalling Professor Teruyo Ojima-Kato and colleagues advanced our understanding of how translation can be modulated from within the ribosome itself. Their discovery that a short tetrapeptide such as SKIK can act in the opposite direction—accelerating translation and mitigating arrest—introduces a new dimension to the biology of nascent chains. It opens the possibility of engineering very short sequence motifs to boost protein yield without the need for bulky fusion tags or extensive redesign of coding sequences. We believe, the implications of this research in biotechnology are immediate. Recombinant proteins with consecutive prolines have long posed problems for production, often forcing researchers to abandon targets or accept poor yields. The finding that a strategically placed SKIK sequence can relieve such stalling provides a practical, inexpensive, and genetically encodable solution. It allows laboratories to rescue otherwise inaccessible proteins without altering their primary structures in ways that could compromise folding or function. By quantifying the kinetic improvements and showing synergy with elongation factor P, the study also offers a rational framework for combining interventions, moving beyond trial-and-error to systematic design. There is also a broader conceptual implication for synthetic biology. Regulatory elements are usually thought of in terms of DNA or RNA—promoters, ribosome binding sites, untranslated regions. This study suggests that small peptide modules, encoded as part of the protein itself, may serve as another layer of control, tuning translation rates dynamically. One can imagine a future where engineered nascent peptide motifs act as programmable switches, enhancing or modulating expression depending on context. This would extend the design space of synthetic circuits and add new versatility to industrial protein production.
Perhaps most importantly, the work calls attention to questions that remain open. How exactly does SKIK exert its effect within the tunnel? Does it change local electrostatics, alter ribosome conformation, or interact transiently with rRNA? Answering these questions will require structural and biophysical approaches, but the foundation laid here ensures they will be pursued. By demonstrating that such a small sequence can exert such a large influence, Professor Teruyo Ojima-Kato and colleagues have not only solved a practical problem but also broadened the conceptual landscape of translation biology.
Reference
Nishikawa Y, Fujikawa R, Nakano H, Kanamori T, Ojima-Kato T. Effect of Translation-Enhancing Nascent SKIK Peptide on the Arrest Peptides Containing Consecutive Proline. ACS Synth Biol. 2024 Dec 20;13(12):3908-3916. doi: 10.1021/acssynbio.4c00221.
 Medicine Innovates Medicine Innovates: Delivering innovations in medicine to the world for better health and prosperity
Medicine Innovates Medicine Innovates: Delivering innovations in medicine to the world for better health and prosperity

